| 일 | 월 | 화 | 수 | 목 | 금 | 토 |
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | ||
| 6 | 7 | 8 | 9 | 10 | 11 | 12 |
| 13 | 14 | 15 | 16 | 17 | 18 | 19 |
| 20 | 21 | 22 | 23 | 24 | 25 | 26 |
| 27 | 28 | 29 | 30 |
- YAI 8기
- GaN
- RCNN
- 자연어처리
- Perception 강의
- Faster RCNN
- VIT
- CS224N
- rl
- 컴퓨터비전
- YAI
- PytorchZeroToAll
- Fast RCNN
- transformer
- 강화학습
- 컴퓨터 비전
- cl
- 연세대학교 인공지능학회
- CNN
- NLP
- YAI 11기
- cv
- YAI 9기
- nerf
- Googlenet
- CS231n
- NLP #자연어 처리 #CS224N #연세대학교 인공지능학회
- 3D
- YAI 10기
- GAN #StyleCLIP #YAI 11기 #연세대학교 인공지능학회
- Today
- Total
연세대 인공지능학회 YAI
[논문 리뷰] U-Net: Convolutional Networks for Biomedical Image Segmentation 본문
[논문 리뷰] U-Net: Convolutional Networks for Biomedical Image Segmentation
_YAI_ 2023. 3. 4. 14:51U-Net: Convolutional Networks for Biomedical Image Segmentation
** YAI 10기 안정우님이 비전논문기초팀에서 작성한 글입니다.
Abstact
Present a network and training strategy that relies on the strong use of data augmentation
- Use available annotated samples more efficiently
The architecture consists of a Contracting path and a symmetric Expanding path
- Contracting path
- Captures content
- Expanding path
- Enables precise localization
- Contracting path
Can be trained end-to-end from very few images
Outperforms the prior best method
- Prior best method: sliding-window conv net
- ISBI challenge for segmentation of neuronal structures in electron microscopic stacks
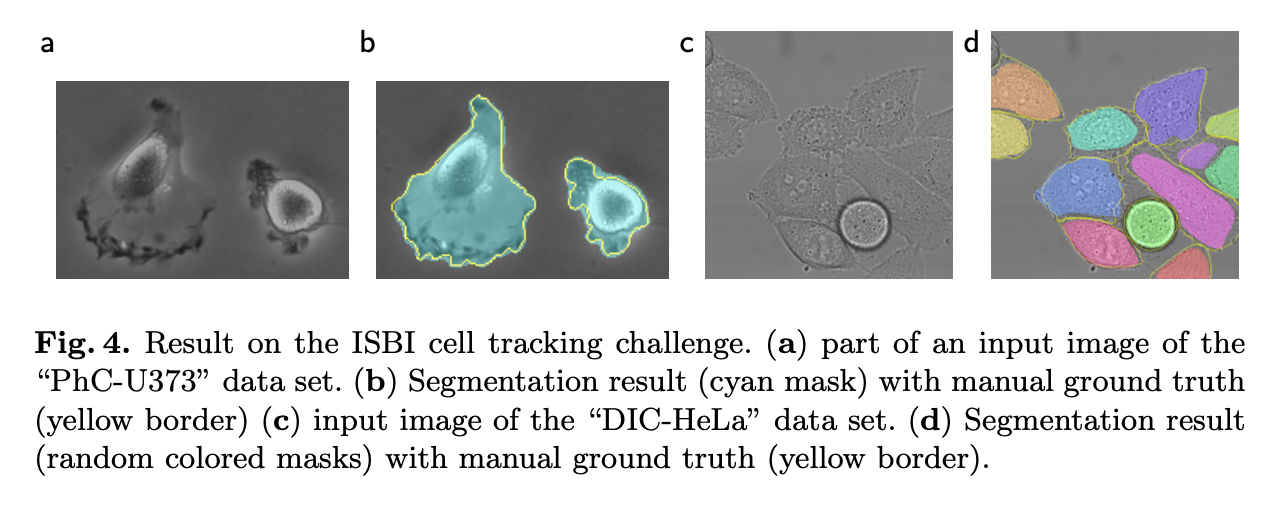
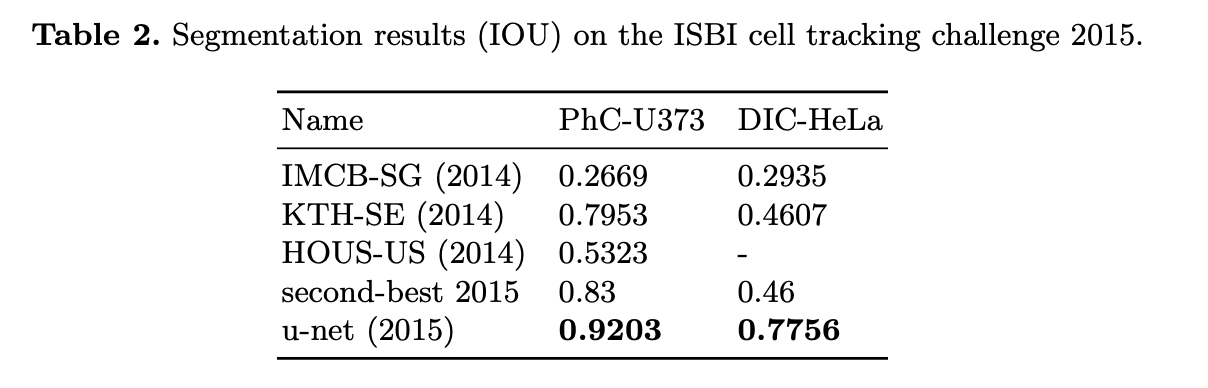
- ISBI cell tracking challenge 2015
Fast Inference
- less than a second on a current GPU
Introduction
Deep Convolutional networks have outperformed the state of the art in many visual recognition tasks
Typical use of conv nets: classification tasks
- Output to an image is a single class label
Segmentation
- in biomedical image processing
- output should include localization
- a class label is supposed to be assigned to each pixel
- in biomedical image processing
Biomedical tasks
- thousands of training images are usually beyond reach
- Ciresan et al.
- trained a network in a sliding-window setup to predict the class label of each pixel by providing a local region (patch) around that pixel as input
- localization available, larger number of training images
- Drawbacks
- Slow
- since the network nust be run separately for each patch, a lot of redundancy due to overlapping patches
- Trade off between localization accuracy and the use of context
- Large patches -> require more max-pooling layers -> reduces localization accuracy
- Small patches -> the network sees only little context
- Slow
Propose new architecture: "U-Net"
- build upon a more elegant architecture, the so-called “fully convolutional network”
- modify, extend this architecture such that it
- works with very few training images
- yields more precise segmentations

- Contracting path
- Expansive path
- Upsampling layers
- increase the resolution
- Upsampling layers
- High resolution features from the contracting path are combined with the upsampled output
- for localization
Upsampling part
- Large number of feature channels -> allows the network to propagate context information to higher resolution layers.
No fully connected layers
- only uses the valid part of each convolution
- the segmentation map only contains the pixels, for which the full context is available in the input image
- allows the seamless segmentation fo arbitrarily large images by an overlap-tile strategy
- Overlap-tile strategy
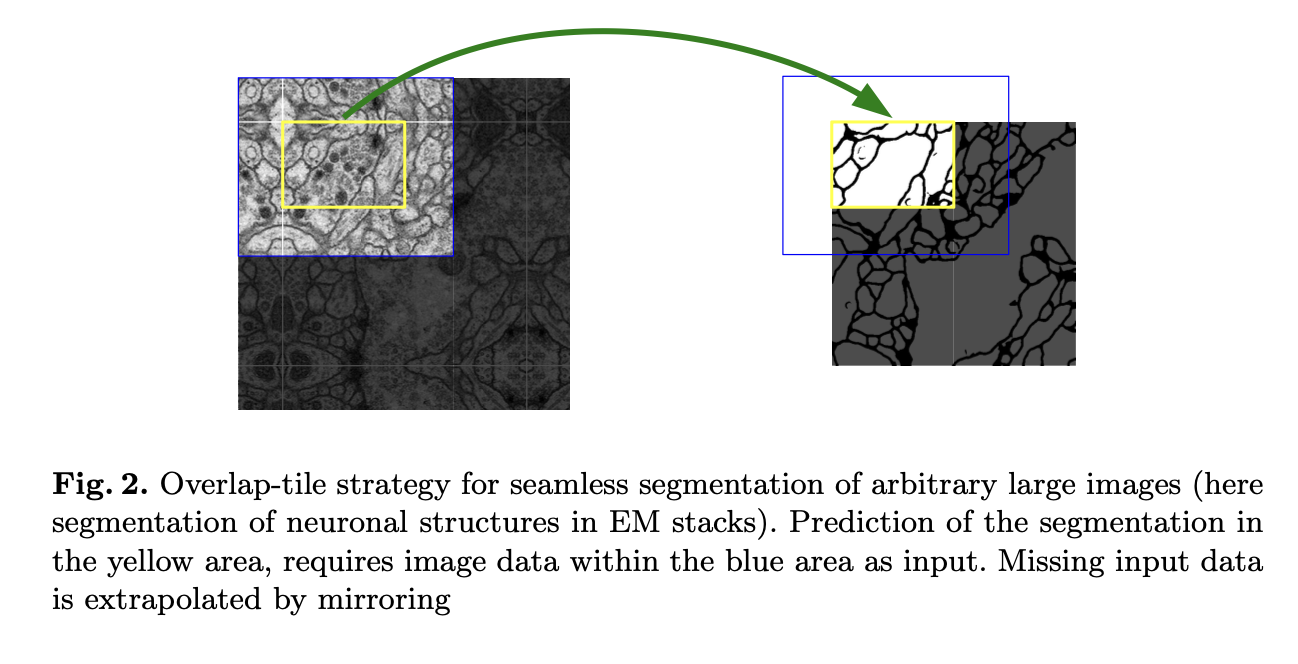
- border region -> extrapolated by mirroring
- Excessive data augmentation
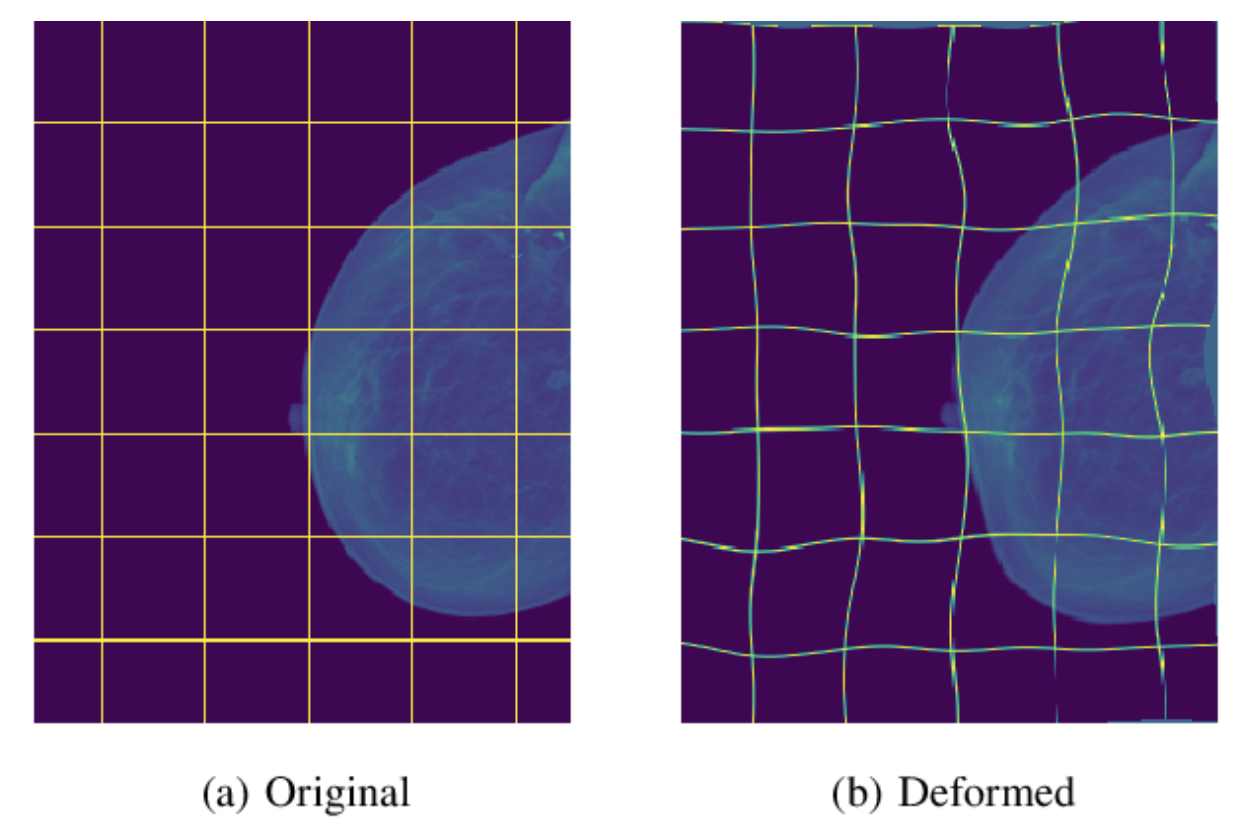
- allows the network to learn invariance to such deformatioins
- particularly important in biomedical segmentation
- deformation used to be the most common variation in tissue and realistic deformations can be simulated efficiently
- Seperation of touching objects
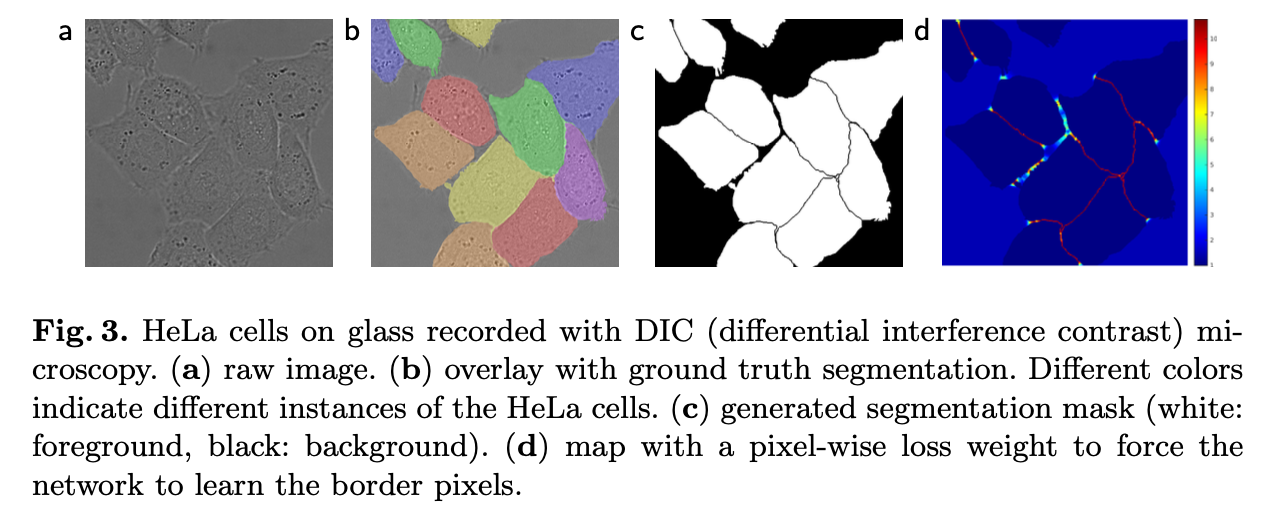
- Weighted Loss
- separating background labels between touching cells obtain a large weight in the loss function
- Weighted Loss
- SOTA Results
Network Architecture

- Contracting path (left side), Expansive path (right side)
- Contracting path
- follows the typical architecture of ConvNet
- two 3x3 convolution (unpadded), each followed by ReLU and 2x2 max-pooling for downsampling
- doubled number of feature channels at each downsampling step
- Expansive path
- 2x2 up-convolution that halves number of feature channels
- concatenation with correspondingly cropped feature map from the contracting path
- two 3x3 convolution, each follwed by ReLU
- 1x1 convolution used to map each 64-component feature vector to the desired number of classes
- total 23 conv layers
Training
- large input tile over large batch size -> reduce the batch size to a single image
- to minimize the overhead and make maximum use of GPU memory
- high momentum (0.99) -> large number of the previously seen training samples determine the update in the current optimization step.
- energy function is computed by a pixel-wise soft-max over the final feature map combined with the cross entropy loss function
- cross entropy
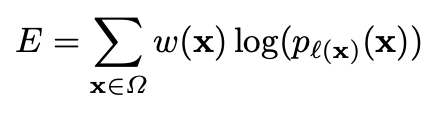

Data Augmentation
- shift, rotation, grayscale, randoms elastic deformations
- Drop-out layers at the end of the contracting path
Experiments

Conclusion
- UNet architecture achieves very good performance on very different biomed- ical segmentation applications
- Thanks to data augmentation with elastic deformations, it only needs very few annotated images and has a very reasonable training time
Discussions
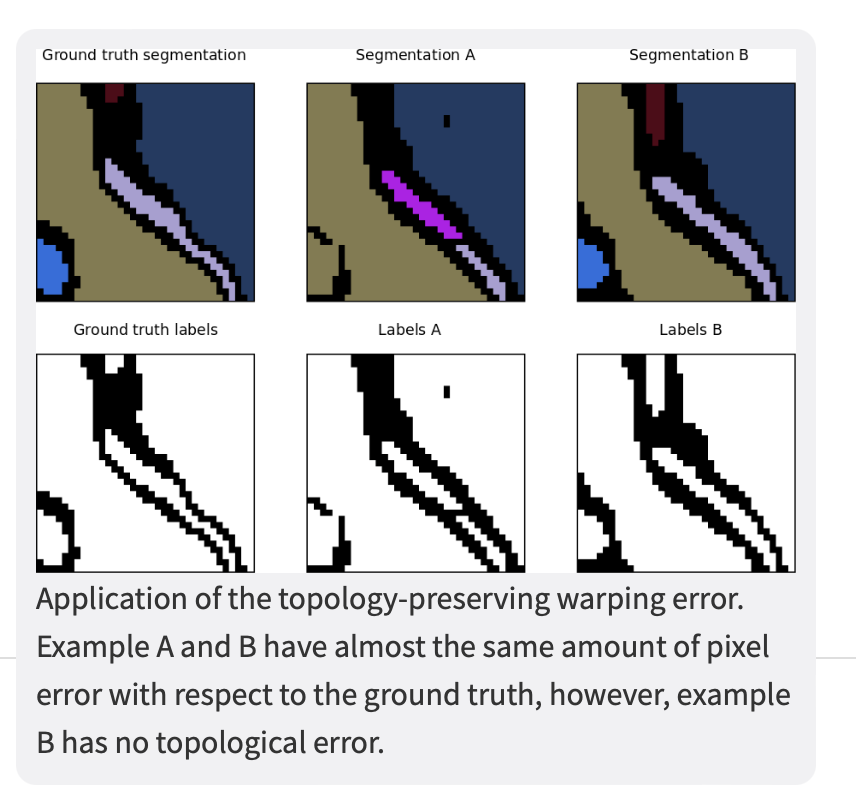
The warping error is a segmentation metric that tolerates disagreements over boundary location, penalizes topological disagreements, and can be used directly as a cost function for learning boundary detection1.
In other words, instead of focusing on the geometric differences (pixel disagreement) between two segmentations, the warping error focuses on the objects and measures the topological error between them.